DDAP Database
Database for T1PKS Pathways
DDAP is a database for type I modular polyketide synthase (PKS), featuring the annotation of the order of ORFs in the biosynthetic pathways.
The order of genes in the biosynthetic pathway of type I polyketides is found to be governed by docking domains (DD) at the N-/C-terminus of the multi-modular polypeptides. There is still much to be learned about DDs and how they affect the assembly line of natural products. To help with the research about docking domains and type I modular PKS (T1PKS), we initiated this database and developed a machine learning based docking domain affinity prediction tool, which predicts the probability of interaction between DDs as well as the most likely pathways for a given sequence of a PKS.
Please cite DDAP database if you would like to use it in your work.
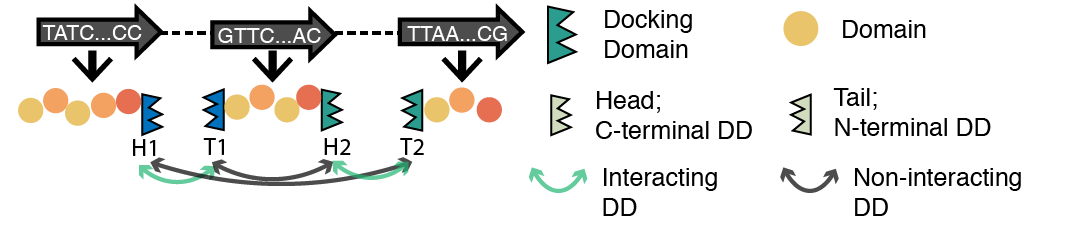
Pathway Prediction Algorithm
We created a machine learning based tool that predicts docking domain affinity as well as the order of ORFs in the biosynthetic pathway of T1PKS. This tool is availabel for downloading at GitHub.
Contribute
You are encouraged to report new T1PKS or pathways of T1PKS. We will update new records regularly. Feedbacks and bug reports are also welcomed. Please leave your comments via GitHub issues. Your support is very much appreciated.
About
Lab of Dr. Arvind Rao
Department of Computational Medicine and Bioinformatics
100 Washtenaw Avenue
University of Michigan
Ann Arbor, MI 48019
Lab of Dr. David H. Sherman
Life Sciences Institute
210 Washtenaw Avenue
University of Michigan
Ann Arbor, MI 48019